Recently, the Ye Xing research team of Pearl River Fisheries Research Institute has completed the mapping and genetic analysis of the genome of largemouth bass, and the research paper "chromosome level genome assembly for the Largemouth Bass Micropterus salmoides provides insights into adaptation to fresh and brackish water" was officially published in the international important academic journal Molecular Biology Ecology resource.
Micropterus salmoides is a member of the genus Micropterus with important ecological and economic significance in the North American aquatic ecosystem, which is naturally distributed in the freshwater basins of North America, but also in the coastal saline and freshwater environment. Since its introduction in 1983, Largemouth Bass has become an important aquaculture fish in China. In 2019, China's aquaculture output will reach 478000 tons. In order to better understand the evolutionary history of largemouth bass, to carry out the research on the genetic basis of its important traits and the development and utilization of germplasm resources, the team carried out the genome research of largemouth bass. By combining pacbio single molecule real-time sequencing and Hi-C technology, the first genome of largemouth bass with high quality chromosome level was obtained. The genome size of largemouth bass is 964mb, contig N50 and scaffold N50 are 1.23mb and 36.48mb respectively. Combined with pacbio RNA sequencing, 23701 genes were annotated.
The haplotype chromosome number of most Perciformes is 24, while that of largemouth bass is only 23. Through comparative genomic analysis, we found that chromosome 1 is a fusion chromosome: chromosome 1 of largemouth bass corresponds to chromosomes 20 and 24 of Lateolabrax japonicus, 18-21 and 24-25 of European bass, respectively. At the same time, a high density telomere element region was found at 24MB in the middle and upper part of chromosome 1 of Largemouth Bass, suggesting that chromosome 1 of largemouth bass consists of two telocentric elements One of the centromeres may be lost or degenerated.
The paper also discussed the molecular mechanism of saltwater adaptation of largemouth bass. Through contraction and expansion analysis, positive selection analysis and transcriptome analysis, 294 genes related to ion regulation were found. Among them, the gene of closure protein of largemouth bass expanded significantly, producing 68 copies of 27 family members, which play an important role in regulating tight junctions between cells, ion and water in and out, and maintaining osmotic stability. The completion of the genome fine mapping of the Largemouth Bass provides an important data base for the protection and improvement of its germplasm resources, the evolution history and the genetic research of important traits.
The journal is one of the top international journals in biochemistry, molecular biology and ecology. It belongs to biology zone 1 (Q1) of JCR division, Chinese Academy of Sciences, with an if of 6.286. The research was supported by the special fund for modern agricultural industrial technology system (cars-46) and the scientific and technological innovation team project of the Academy of water sciences. The Pearl River Fisheries Research Institute of Chinese Academy of Fishery Sciences is the first unit to complete the paper. Dr. Sun Chengfei and Dr. Li Jia are the co first authors of the paper. Researcher Ye Xing and researcher Shi Qiong are the co corresponding authors of the paper.
Article link: https://onlinelibrary.wiley.com/doi/epdf/10.1111/1755-0998.13256
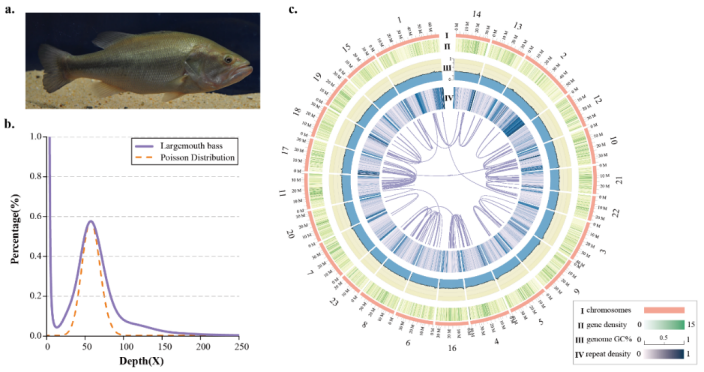
Fig. 1The chromosome-level genome assembly and circos atlas of the largemouth bass.
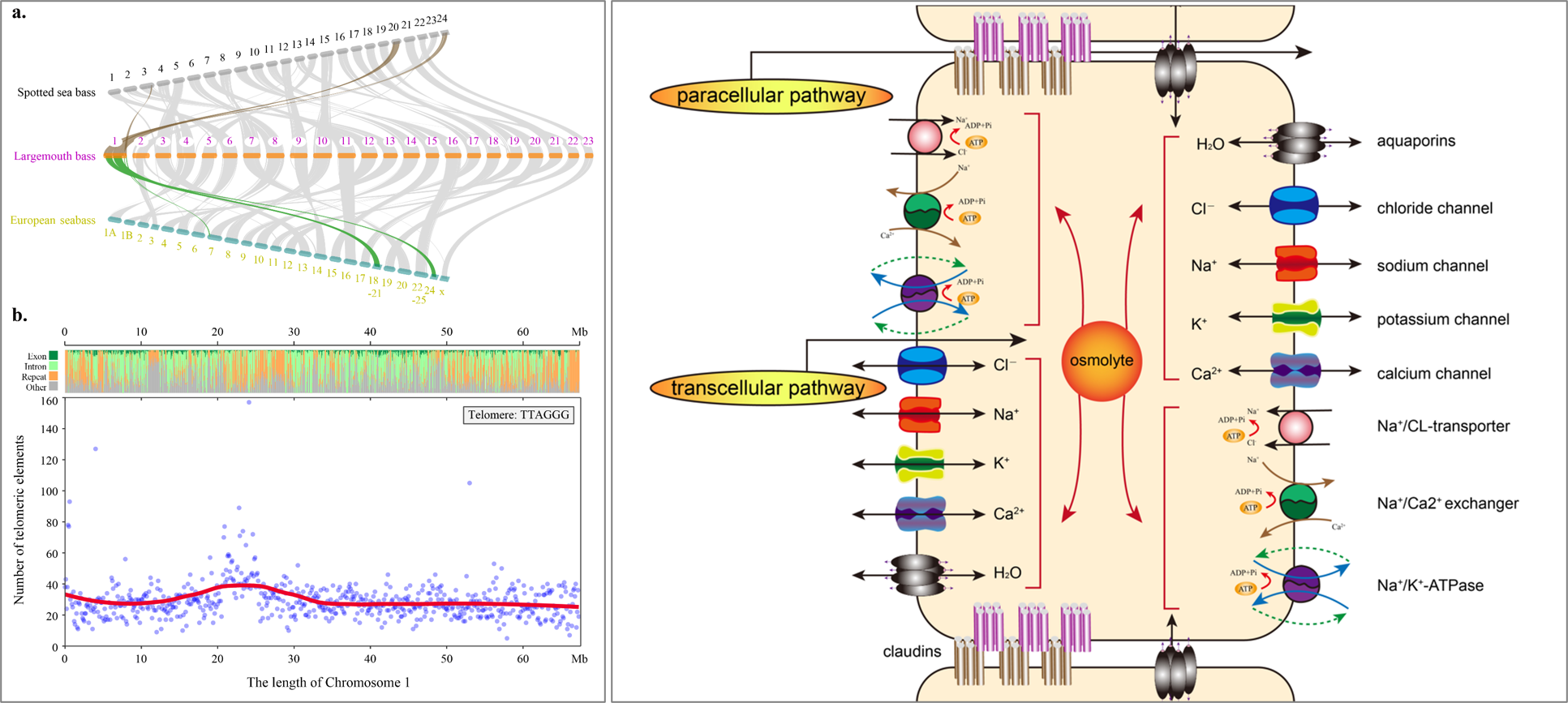
Fig. 2Chromosomal fusion in the Chr1(Left)and the proposed gene network map for ion regulation in LMB(Right)

